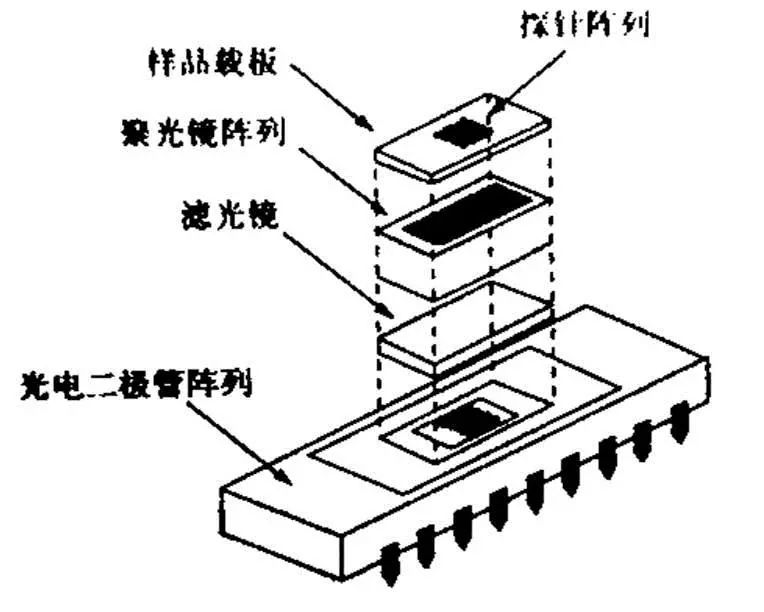
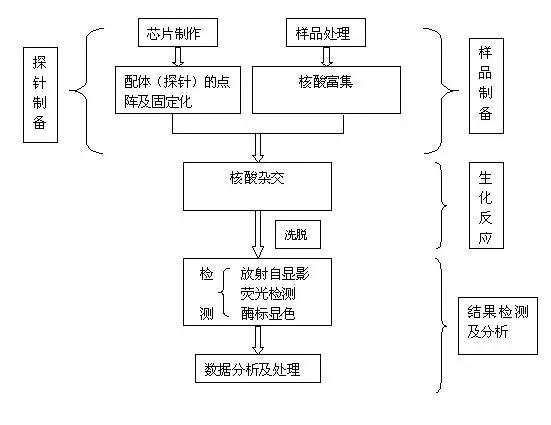
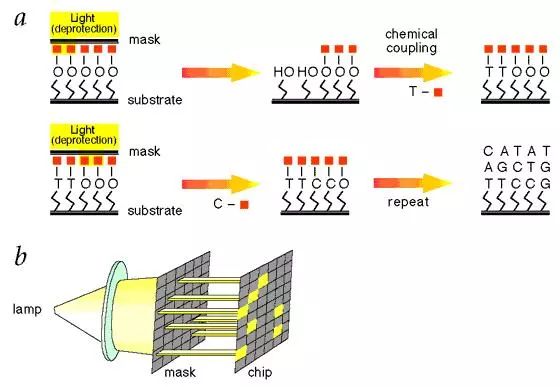
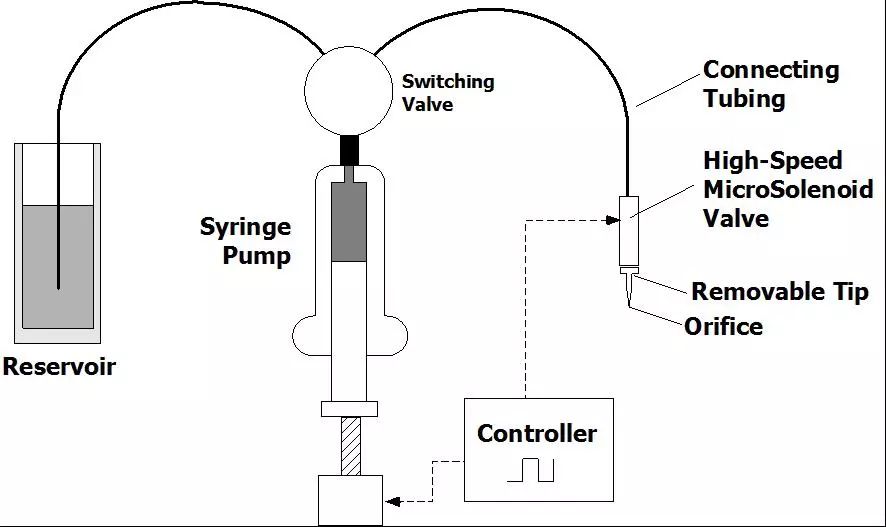
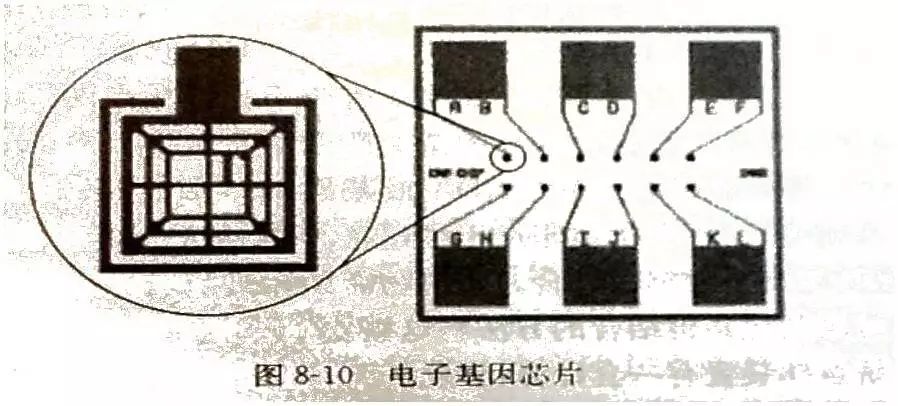
Gene chip technology is a kind of biochip. It is a high-tech emerging in the field of life sciences. It integrates advanced technologies such as microelectronics manufacturing technology, laser scanning technology, molecular biology, physics and chemistry. Biochip Biochips refer to the orderly consolidation of thousands of target molecules (such as DNA, RNA or proteins, etc.) onto smaller supports (such as glass sheets, silicon wafers, nylon membranes, etc.). The dense molecules are arranged, and the labeled sample is then hybridized with the target molecule on the support. After elution and laser scanning, the resulting signal is automatically analyzed using a computer. This method not only saves reagents and samples, but also saves a lot of manpower, material resources and time, making detection more rapid, accurate and sensitive. It is currently the most efficient, most sensitive and most promising technology in biological detection. Biochips can be classified into gene chips, protein chips, tissue chips, and chip labs depending on the type of target molecules immobilized on the support. At present, the technology is mature and most widely used is the gene chip technology, which has extensive applications in genome expression analysis, drug screening, gene expression and functional research of simulated organisms, genetic disease genetic diagnosis, and diagnosis of pathogenic microorganisms. , is an efficient and large-scale acquisition of relevant biological information an important means. Gene chip Gene chips, also known as DNA microarrays, are a type of biochip. The principle of the gene chip is originally derived from the molecular hybridization of nucleic acids. That is, a nucleic acid probe of known sequence is used to perform hybridization detection on a nucleic acid sequence of an unknown sequence. The DNA chip technology is actually a large-scale integrated solid-phase hybridization. Refers to the in-situ synthesis of oligonucleotides on a solid support or the direct solidification of a large number of pre-prepared DNA probes to the surface of the support in a microprinting manner, followed by hybridization with the labeled sample. . Through computer detection and analysis of the hybridization signal, the genetic information of the sample (gene sequence and expression information) was obtained. Because the computer silicon chip is often used as a solid support, it is called a DNA chip. The gene chip uses a large number of specific oligonucleotide fragments or gene fragments as probes, and is regularly fixed on a solid support such as a silicon wafer, a glass sheet, a plastic sheet, or a nylon substrate combined with an electro-optical measurement device to form a two-dimensional array. The array, the gene of the labeled sample to be tested, hybridizes on the principle of base-pair pairing to detect a specific gene. The gene probe uses a hydrogen bond between the complementary bases of the ribose duplex to form a stable double bond structure, and the sample is detected by measuring the photoelectric signal on the target gene, so that the gene chip technology becomes an efficient large-scale acquisition. The important means of biological information. Gene chip technology principle 1. Massive collection and purification of DNA probes. Gene chip probe preparation methods can be based on the design of specific PCR primers to specifically expand the gene, or it can be to establish a homogenized cDNA library. Screening, amplification; 2, the purified probe is fixed on the substrate, first to the substrate (mainly glass) is subjected to special chemical treatment, the glass sheet aldehyde or amination, and then pass the purified probe Micrographic printing or jetting on the substrate, and then printing the finished glass sheet for post-processing, such as hydration, heating or UV crosslinking; 3. The marking of the sample, marking method is generally used by reverse transcription or random primer extension method; 4. Scanning of hybridized chips, acquisition of image processing, and data analysis. Electronic chip (left) vs. gene chip (right) Gene chip features 1. High-throughput, multi-parameter synchronous analysis. At present, the gene chip manufacturing process can achieve a fixed number of tens to hundreds of thousands of probes on a 1 cm2 carrier plane, and can synchronously detect and analyze a large number of related genes in the sample, and even the entire genome and information. 2, rapid and automatic analysis. Under certain conditions, the target gene fragment in the sample is simultaneously hybridized with multiple probes of the chip, and the hybridization signal and analysis processing data are measured using a scanning instrument. Thus, the speed and efficiency of the measurement work are fundamentally improved, and the intensity and difficulty of the measurement work are also greatly reduced. 3, high accuracy analysis. Because each point on the chip, that is, each probe can be accurately located and sited, and each probe can be accurately designed and prepared, different target genes and different states of the same target gene can be accurately detected and The difference in one base. 4, high precision analysis. The unification of precision and detection reagents and methods for commercial chip production ensures the high precision and reproducibility of the chip detection to a certain extent, so that the test results between batches and even different laboratories can be effectively compared. Pair and analyze. 5, high sensitivity analysis. The gene chip selects a vector that does not easily produce a diffusion effect. The hybridization points of the probe and the sample target gene are very concentrated. In addition, the amplification of the target gene before hybridization and the expansion of the detection signal after hybridization greatly improve the detection sensitivity. Up to one copy of the target gene can be detected in one cell, so that the amount of sample required for detection is greatly reduced, and generally only 10 to 20 μL of the sample is required. Gene chip classification Gene chip types are numerous and can be classified according to different classification methods. Generally, they can be classified into the following types: 1. According to the different kinds of DNA added on the carrier, the gene chip can be divided into two kinds: oligonucleotide chip and cDNA chip: Oligonucleotide chips are generally fixed to the carrier by means of in-situ synthesis and have a high degree of density. It can synthesize the advantages of any series of oligonucleotides, and is suitable for DNA sequence determination, mutation detection, SNP analysis, etc. The disadvantage is that the length of synthetic oligonucleotides is limited, so the specificity is poor, and as the length increases, The synthesis error rate increases. Oligonucleotide arrays can also be prepared by pre-synthesizing spots, but the fixation rate is not as high as that of cDNA arrays. Oligonucleotide arrays are mainly used for point mutation detection and sequencing, and can also be used as expression profile studies. The cDNA chip is a microarray densely packed and solidified micro-sized cDNA fragments in a matrix such as glass. Although the spot density of the cDNA is not as high as that of the in-situ synthesized oligonucleotide chip, it is much higher than that of a conventional vector. The biggest advantage of cDNA microarrays is that the specificity of target gene detection is very good. It is mainly used for expression profiling studies. 2, according to the carrier material classification: carrier materials can be divided into two kinds of inorganic materials and organic materials, inorganic materials are glass, silicon, ceramics, organic materials from the organic film, gel and so on. The membrane chip is mainly composed of a nylon membrane. Its array density is relatively low, and the amount of probe used is large. The detection method is mainly a method using radioactive isotopes. The result of the detection is a monochromatic result. The glass-based chip has a high density of arrays, uses fewer probes, and has a variety of detection methods. The result is a color result. Compared with the membrane chip, the resolution is higher and the analysis is more flexible. More powerful. 3, according to the different methods of spotting can be divided into in situ synthesis chip, micro-matrix chip, electro-positioning chip three. Gene chip preparation technology Traditional preparation techniques Because there are many types of chips, the preparation methods are not the same. The traditional preparation methods can basically be divided into two categories: one is in-situ synthesis and the other is direct spotting. In situ synthesis is used for oligoamino acids, direct spotting is mostly used for large fragment DNA, and sometimes also for oligonucleotides or even mRNA. In situ synthesis includes photolithography and piezoelectric printing. 1. Original photolithography Its use of solid-phase chemistry, photo-sensitive protective groups, and photolithographic techniques yields well-defined, highly diverse sets of compounds. The first step in the synthesis is to use light irradiation to deprotect the hydroxyl groups on the solid surface, and then the solid surface is contacted with a photoprotective group-protected phosphoramidite-activated base monomer to connect a single nucleotide monomer. The synthesis occurs only where the protective group is removed. This process is repeated until the synthesis is completed. The biggest advantage of this method is that in a smaller area, a large number of different probes can be manufactured. However, this preparation method needs pre-selection design, manufactures a series of masks, the cost is high, and the light deprotecting method is used in the manufacturing process. When the masking aperture is small, light diffraction phenomenon occurs, which restricts the further increase of the probe density. 2. Original print synthesis This principle is similar to ink printing, but there are multiple chip print heads and ink cartridges. The ink cartridges contain four bases of liquid rather than carbon powder. The print head can move across the chip and be based on different positions on the chip. Spot probe sequences are needed to print specific bases on specific locations on the chip. The chemistry used in this technique is consistent with the traditional solid-phase DNA synthesis and therefore does not require specially prepared chemical reagents. 3. In-situ synthesis of molecular seals The synthesis principle is similar to the traditional seal, and the surface is made into an uneven surface according to the requirement of array synthesis. According to this, different nucleic acid or polypeptide synthesis reagents are printed on a specific site of the chip base, and then the synthesis reaction is performed. 4. Spot method Compared with the in-situ synthesis method, the spotting method is relatively simple. Only a sample of an oligonucleotide or cDNA prepared in advance is spotted on a glass sheet or other material that has been specially processed in situ by an automatic spotting device. That is, the samples can be purified in advance, and the cross-linking manners are various. Furthermore, by adjusting the concentration of the probes, the hybridization signals of the probes composed of different bases can be made uniform, and the researchers can easily design and prepare a gene chip that meets their own needs. However, in the preparation process of the chip, the waste of the sample is serious, and the chemical modification of the oligonucleotide also increases the synthesis cost, and a large amount of sample needs to be stored before the chip is prepared. New preparation technology Microelectronics chip Using the lithography technology commonly used in the microelectronics industry, the chip is designed to be built on a silicon/silicon dioxide base material, as shown in Figure 2, and thermally oxidized to make an array of 1mm x 1mm, each array containing multiple micro Electrodes, a sample cell was prepared by silicon oxide deposition and etching on each electrode. The streptavidin-linked agarose is coated on the electrode, and the biotin-labeled probe can be bound to a specific electrode under the action of an electric field. The biggest feature of the electronic chip is the fast hybridization speed, which can greatly shorten the analysis time, but the preparation is complicated and the cost is high. 2. Three-dimensional biochip This chip technology mainly uses functionalized polyacrylamide gel pieces as a matrix to immobilize oligoamino acids. The usual preparation method is to polymerize the active group-containing substance or acrylamide derivative and acrylamide monomer on a glass plate and mechanically cut out the three-dimensional gel micro-blocks so that there are 10,000 tiny polyethylenes on each glass plate. Amide gel strips, each gel strip can be used for the analysis of target DNA, RNA or protein, lithography or laser evaporation to remove the gel between the gel blocks, and then with active groups (amino, aldehyde, etc.) The DNA spot was added to the gel for cross-linking and the DNA sample was transferred to the gel block. 3. Flow-through chip That is, grid-like microchannels are fabricated on the chip substrate, and specific oligoamino acid probes are designed and synthesized to be integrated into specific regions of the microchannel within the chip. The DNA or RNA is separated from the sample to be detected and labeled with fluorescence. The sample then flows through the chip. The immobilized oligo-amino acid probe captures the complementary nucleic acid, and the signal detection system analyzes the result. Its characteristics are high sensitivity, fast speed, and low price. The current major gene chip technologies Optically guided in situ synthesis of oligonucleotide microarrays Developed and mastered by Affymetrix, Affymetrix uses photolithography technology combined with light-guided in situ oligonucleotide synthesis to produce DNA chips. The production process is very similar to that of electronic chips. Gene chips produced using this technique can achieve a microprobe array density of 1×10 6 /cm 2 , allowing millions of oligonucleotide probes to be arrayed on a piece of 1cm square. The in-situ synthesis method is mainly a light-directed synthesis technique. It can be used not only for the synthesis of oligonucleotides but also for the synthesis of oligopeptide molecules. Photo-guided polymerization is a combination of photolithography and traditional solid-phase synthesis of nucleic acids and peptides. Semiconductor technology has used photographic plate technology to fabricate microelectronic circuits on semiconductor wafers. Solid-phase synthesis technology is a commonly used method for the synthesis of polypeptides and nucleic acids, and the technology is mature and automated. The combination of the two provides a quick way to synthesize high-density nucleic acid probes and short peptide arrays. Affymetrix has already marketed diagnostic microarray products. They can be divided into three categories according to their uses: gene expression chip, gene polymorphism analysis chip and disease diagnosis chip, gene expression analysis chip and gene polymorphism analysis chip. For research institutions and biopharmaceutical companies, they can be used to find new genes, gene sequencing, disease gene research, genetic pharmaceutical research, new drug screening, and many other fields. Affymetrix mainly manufactures general-purpose oligonucleotide chips; disease diagnosis chips are mainly used In medical clinical diagnosis, including various genetic diseases and tumors, Affymetrix currently produces three commercially available diagnostic chips, namely the p53 gene mutation diagnostic chip, the HIV gene mutation diagnostic chip, and the cytochrome P450 gene mutation diagnostic chip. Microelectronics chips Nanogen has developed a multi-site electronically controlled array of microelectronic microarrays containing independently addressable detection regions. The matrix is ​​entirely made up of silicon, germanium, and basic semiconductor materials, on which 25-400 micro-platinum electrode sites are constructed. Points can be controlled by computers independently or in combination. Whether in chip manufacturing or on-chip detection, nucleic acids can be combined by electric field changes similar to the microelectrode, and the “electronic stringency†parameter is introduced so that the chip detection can control the hybridization process through the target, probe sequence characteristics and user requirements. Strictness. This microelectronic gene chip has the following advantages: 1. The electric field localization process can selectively transport charged DNA molecules, change the positive, negative, and strong electric fields of each microelectrode site, and can accurately and effectively regulate the nucleic acid on the surface of the chip, and can bind the nucleic acid to the microelectrode sites. On the other hand, the nucleic acids can also be transported. 2. The rate of DNA hybridization can be accelerated by changing the electric field. By introducing a positive electric field, the binding rate of the nucleic acid to be measured to a known probe can be greatly accelerated, and the hybridization reaction time can be reduced, compared with several hours of an ordinary "passive" hybridization reaction. This "active" hybridization reaction can be accomplished in just a few seconds. In addition, the change of the electric field can effectively remove the unbound free molecules and reduce the interference of the unbound fluorescence signal. 3. The electronic stringency can effectively control the degree of mismatch in the hybridization process, the degree of hybridization mismatch, and different electrical fields can be used to meet different electronic stringency requirements for different requirements. This can be very stringent for nucleic acid hybridization. Flexible control, which allows very accurate SNP detection. Micro spotting technology At present, most of the companies that produce microarrays use this method. They use advanced and more micro-spotting techniques to spot even more tiny probes. The on-chip probe produced by this method is not limited by the type of probe molecular size and can flexibly and flexibly produce a chip according to the user's requirements. Since there are production and testing instruments sold at the same time, the supporters can make corresponding chips according to their own needs, and the price is low, so there will be a certain market in the near future. There are many companies that produce such equipment, such as Genomicsolutions in the United States, BioRobotics in the United Kingdom, Cartesian in the United States, and Engineering in Canada. For the microchip produced by the micro spotting technology, the instrument composition can be divided into spotting instruments, hybridization devices, detection instruments, and analysis instruments. The spotting instrument is advanced to determine the density of the probes on the chip and the degree of firmness of the bonding chip. The density of the probe is an important indicator. To achieve a very high density of probe arrays is the goal that many chip production companies dream of, but the specific spotting density is determined according to the user's purpose, but also to consider the subsequent hybridization. And testing process. There are several more important indicators for measuring the spotting device, such as the overall design of the instrument, the diversity of functions, the diversity of the chip matrix, the stability of the spotting, the spotting speed, the spotting density, and the like. Dotted arrays generally use solid or hollow spotted needles. There are two ways of spot printing: inkjet printing and contact printing. Currently, there are two types of non-contact point-of-spot technology for DNA spotting. One is a piezoelectric technology that uses a piezoelectric crystal to eject liquid from a hole. The droplet size is generally 50-500 pl. This is a syringe-solenoid technology. This technique is precisely controlled by the combination of high-resolution syringe pumps and micro- solenoid valves. Testing equipment is also an important constraint. If the resolution of the testing instrument is not high, then even a very high density chip will not be useful if the spotting instrument is used. For high density chips, laser confocal microscopes and high performance are usually used. Cooling the CCD has both advantages and disadvantages and must be comprehensively measured according to requirements. The chromogenic and analytical methods for color development and analysis are mainly fluorometric methods. Mass spectrometry, chemiluminescence, and optical fiber methods are currently being developed. Taking the fluorescence method as an example, the current major detection methods are laser confocal microscopy and high-performance cooling CCDs to facilitate the quantitative analysis of the fluorescence intensity at each site of the high-density probe array. Because the intensity of the fluorescence signal generated when the probe is perfectly matched to the sample is 5-35 times that of a single or two mismatched base probes, accurate determination of fluorescence signal intensity is the basis for achieving detection specificity. Analytical instruments are only high-performance computers in terms of hardware, but the most important one is analytical software. If only simple tests or scientific experiments are performed, the genes to be analyzed in the samples to be tested are rarely very simple and intuitive observations are used. It can be concluded that, for a large number of genetic analysis or the use of clinical inspectors, comprehensive and intelligent analysis software is required to assist, and this also requires consideration of software upgrades. Other technologies Mainly the United States NIH, Caliper and Orchidbio, etc., Orchidbio company developed a capillary microfluidic pump chip, integrates 144 micro-chambers on the 2-inch side of the chip, respectively, from the inflow hole, reaction chamber, circulation tube and Outflow hole composition, this chip can not only be used for genetic diagnosis and analysis, but also for synthetic chemistry, the use of chip microfinger structure, Caliper's chip can be used as a cell sorter, able to use the blood cell volume and deformability Equivalent features make it easy to separate red blood cells from white blood cells. NIH has developed a microchip reactor that can quickly complete a series of biochemical reactions. Gene chip technology applications Research field At the end of 1998, the American Science Promotion Association listed the gene chip technology as one of the ten major progresses in natural science in 1998, which shows its significance in the history of science. It shows tremendous power with its ability to analyze thousands of genomic information simultaneously, quickly, and accurately. These applications include gene expression detection, mutation detection, genomic polymorphism analysis, gene library mapping, and hybridization sequencing. Using gene chips to detect changes in gene expression can save a lot of manpower, material resources and financial resources. In the past, scientists had to repeat a large number of experiments to observe changes in multiple genes, if the traditional method of studying thousands of genes in the cell The change is almost unthinkable because it is necessary to extract the nucleic acid that first extracts the cells, but also enough to meet the needs of the Northern hybridization, then label each probe and perform the hybridization assay separately. The use of the gene chip can reduce the workload by tens of thousands of times. The function of 6,000 genes in the yeast can be simultaneously detected using the gene chip, while the research team led by Patrick Brown of Stanford University has successfully detected human fibroblasts. The expression of 8,600 genes changed. GeneChip Black can be used for gene sequencing. At present, the United States Human Genome Project is developing this technology to replace the current automatic sequencing. Compared with the existing manual sequencing and automated sequencing, gene chip sequencing can save a lot of reagent and instrument loss . In the research of gene expression detection, people have successfully studied the genome expression of various organisms including Arabidopsis thaliana, yeast and human. It has been proved that the gene chip technology can also be applied to the detection of nucleic acid mutations and the analysis of genomic polymorphisms, mutation detection that is 98% identical to conventional sequencing results, and identification and mapping of single nucleotide polymorphisms in the human genome. Classification, human mitochondrial genome polymorphism, etc. Combining the biosensor and the chip technology, it has been proved that the single-base mutation of the gene can be detected by changing the electric field intensity of the probe array region, and the yeast genome can be mapped by determining the order of overlapping clones. Hybridization sequencing is another important application of gene chip technology. The sequencing technology is theoretically an efficient and feasible method of sequencing, but the sequence of the target nucleic acid molecule must be deduced through the hybridization of a large number of overlapping sequence probes with the target molecule, so a large number of probes need to be produced. Gene chip technology can easily synthesize and immobilize a large number of nucleic acid molecules, so the advent of it will undoubtedly provide practical possibilities for hybridization sequencing. Biopharmaceutical field Major pharmaceutical companies and biotech companies will use gene chip discovery to screen for new drugs. The adoption of gene chip technology can greatly speed up the progress of the human genome project, such as gene sequencing, gene expression detection, and new genetic markers such as SNP mapping, which is to find new functional genes and find new drug targets. It is of great significance to develop new gene drugs. The use of a gene chip allows for the detection of gene expression changes under different species, different tissues, different diseases, and different processing conditions, which enables the development of diagnostic kits with different uses. New drugs must pass human safety experiments at the experimental stage, and they must observe the effects of drugs on human gene expression. Since they do not know that drugs work for that kind of genes, they must perform all known or a certain range of gene expression. Detection, the use of gene chips can complete this task quickly and accurately, the United States Tularick company has developed a new drug that can significantly reduce low-density lipoprotein - a substance that can cause hardening of the arteries, followed by Tularick's scientific research The researchers used Syntini's gene chip to study the effect of the new drug on human cell gene expression and found that it could significantly alter the gene expression profile of the cell, which is very similar to another toxic reaction. Tularick had to stop the drug. R & D, this company saves a lot of investment. Medical diagnosis 1. In eugenics, we now know that there are more than 600 genetic diseases related to genes. Women using DNA chips for genetic diagnosis early in pregnancy can avoid many genetic diseases. 2. In terms of disease diagnosis, since most diseases are related to genes and are often associated with multiple genes, DNA chips can be used to find the correlation between genes and diseases, thus developing corresponding drugs and proposing new therapeutic methods. The high density of DNA chips and the advantages of parallel processors not only make multi-gene analysis possible, but also ensure that diagnostics are efficient, inexpensive, fast, and simple. 3. Genome matching for organ transplantation, tissue transplantation, and cell transplantation, such as HLA typing. 4. Diagnosis of pathogens, such as identification of bacteria and viruses, identification of drug resistance genes. 5. In terms of the environmental impact on the human body, it is known that pollen allergies and other human reactions to the environment are related to genes. The comprehensive monitoring of more than 200 genes related to environmental pollution will have important implications for ecological environment control and human health. 6. In forensic science, the DNA chip goes further than previous DNA fingerprinting. It can not only do gene identification, but also depict the facial appearance of living organisms through the life information contained in the DNA. This test is often used after a disaster to identify the identity of the body and to identify the blood relationship between parents and children. This shows that using DNA chips can quickly and efficiently obtain unprecedented scale of life information. This feature will make DNA chip technology a revolutionary new method and new tool for scientific exploration and medical diagnosis in the future. Suizhou simi intelligent technology development co., LTD , https://www.msmsmart.com